3D Local in Vivo Environment (LivE) Imaging
This research focuses on expansion and optimisation of a novel methodological platform known as LivE imaging with the ultimate aim of using the methodology to determine the significance of osteocyte proteins in healthy and pathological bone remodelling. The LivE imaging platform uses a variety of steps, to allow the mechanical and remodelling environment within 2D sections of bone tissue to be linked with osteocyte protein expression patterns. Previously this link has remained difficult to establish, as osteocytes are embedded within bone tissue making their analysis difficult.
The LivE imaging methodology includes the following aspects; 1. In vivo micro-computed tomography is used to track the remodelling activity of a mouse vertebra. 2. The vertebra is reconstructed in 3D and areas of formation, resorption and quiescence are identified within the reconstruction. 3. Finite element analysis is used to identify the strain environment within the reconstruction. 4. Immunohistochemical staining is used to identify protein expression in 2D sections of osteocytes. 5. The histological section is processed and registered within the 3D reconstruction of the vertebra. 6. The remodelling, strain and cell staining data is processed using software developed in house to connect the location of individual cells, their protein expression and the local environment.
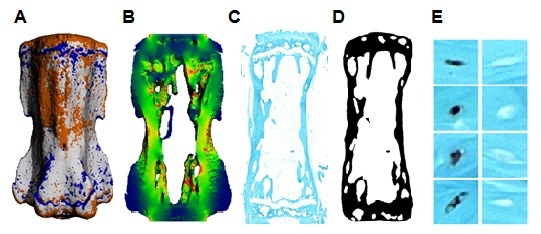
Through the application of LivE imaging it has been possible to associate the protein sclerostin with bone resorption activity in 2D sections. However, osteocytes form a 3D network throughout bone tissues. Therefore the specific aim of this project has been to expand the LivE imaging method to incorporate 3D data sets, i.e. serial sections of vertebral tissues. In addition, various steps within the methodological platform are being investigated and optimised to increase the robustness of the method.
In future, the 3D LivE imaging platform can be used to investigate the effects of osteocyte proteins on anabolic and catabolic bone remodelling. This has major implications for our understanding of bone diseases such as osteoporosis. Therefore, the method has clinical significance, as it will be possible to use the platform to identify osteoporotic targets.