In Vivo Single-Cell Transcriptomics in Bone
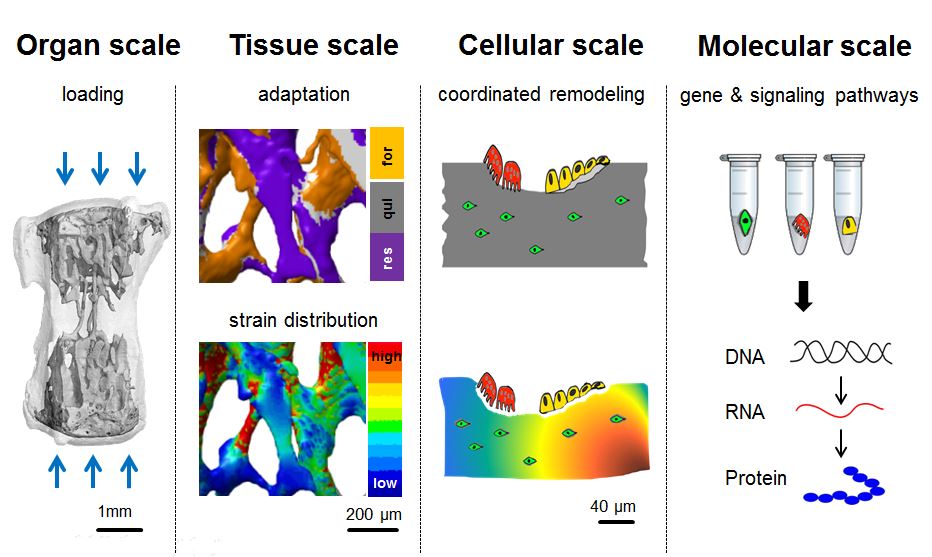
Bone is a dynamic tissue that adapts its architecture to its mechanical needs. Forces acting on bone at the organ scale stimulate bone formation at the tissue scale by affecting gene expression and signaling pathways at the cellular scale. Hence, the understanding of load-induced bone remodeling requires an experimental approach that spans the different scales. However, the diversity of cell types present in bone together with the intricate interplay between them makes it difficult to assess the functional role of each one of them separately. Furthermore, as bone remodeling occurs asynchronously at many sites throughout the skeleton, the remodeling activities have to be assessed locally.
Global gene expression assays derived from in vivo models for load-induced bone remodeling have identified a number of candidate genes and revealed potential load-regulated pathways. The techniques that have facilitated this are cell harvesting, i.e. the analysis of gene expression from cell populations isolated from loaded bone. However, these bulk assays report gene expression averaged over tens of thousands of cells, thereby concealing potentially important signals such as biologically relevant cell-to-cell variability. More importantly however, these techniques require the mechanical or enzymatic disaggregation of the entire tissue, which unavoidably causes loss of spatial information. Considering that each of these cells resides in a different micro-environment characterized by different levels of mechanical strain and local remodeling activity, preserving the spatial information is of paramount importance for successful deciphering of the molecular pathways involved in these processes.
By combining RNA sequencing of single cells isolated by laser-capture microdissection with micro-finite element analysis based on time-lapsed in vivo micro-computed tomography, we propose a systems level approach to allow spatially resolved transcriptomic profiling of individual cells from mechanically loaded bone. This approach will not only provide new insights into bone mechanobiology, but furthermore, will represent a generally applicable methodology which will enable cells and their entire transcriptome to be mapped in three dimensions in in vivo animal models, thus making it possible to investigate how cellular processes are choreographed in space and time in whole organs and even organisms.